In this example we show how to generate a mesh with toroidally non-continuous conductors for the LTX-β device using TokaMakers built in mesh generation.
- Warning
- Toroidally non-continuous conducting regions are still in development. Please be careful when using this feature and report any issues.
- Note
- Running this example requires the h5py python packages, which is installable using
pipor other standard methods.
Load TokaMaker library
To load the TokaMaker python module we need to tell python where to the module is located. This can be done either through the PYTHONPATH environment variable or using within a script using sys.path.append() as below, where we look for the environement variable OFT_ROOTPATH to provide the path to where the OpenFUSIONToolkit is installed (/Applications/OFT on macOS).
For meshing we will use the gs_Domain class to build a 2D triangular grid suitable for Grad-Shafranov equilibria. This class uses the triangle code through a simple internal python wrapper within OFT.
Build mesh
Set mesh resolution for each region
First we define some target sizes to set the resolution in out grid. These variables will be used later and represent the target edge size within a given region, where units are in meters. In this case we are using a fairly coarse resolution of 2 cm in the plasma region and 5 cm in the vacuum region. Note that when setting up a new machine these values will need to scale with the overall size of the device/domain. Additionally, one should perform a convergence study, by increasing resolution (decreasing target size) by at least a factor of two in all regions to ensure the results are not sensitive to your choice of grid size.
Load geometry information
The geometry information (eg. bounding curves for vacuum vessels) are now loaded from a JSON file. This JSON file contains the following:
limiter: A contour of R,Z points defining the limiter surface (inner side of shell, bridged at the midplane at the back side)shell: A contour of R,Z points defining the a single shell segment (upper)vv: A set of contours defining the vacuum vessel. Multiple regions are used to specify different resistivity where ports are placed.coils: A dictionary of R,Z,W,H values defining the PF coils as rectangles in the poloidal cross-section
A JSON is a good way to store common device information that will be reused many times. For simple geometries, testing, or generative usage this can be created directly in the code.
Define regions and attributes
We now create and define the various logical mesh regions. In the LTX-β case we have 5 region groups:
air: The region outside the plasma limiter. Note that this is one region as this model does not have a connection on the inboard side so the VV is not airtight.plasma: The region inside the limiter where the plasma will existshellU,shellL: The upper and lower shellvv1,vv2,...: The individual sub-regions that make up the vacuum vesselOH1,...: Each of the 17 coils in LTX (5 OH, 12 PF)
For each region you can provide a target size and one of four region types:
plasma: The region where the plasma can exist and the classic Grad-Shafranov equation with \(F*F'\) and \(P'\) are allowed. There can only be one region of this typevacuum: A region where not current can flow and \(\nabla^* \psi = 0\) is solvedboundary: A special case of thevacuumregion, which forms the outer boundary of the computational domain. A region of this type is required if more than one region is specifiedconductor: A region where toroidal current can flow passively (no externally applied voltage). For this type of region the resistivity should be specified with the argumentetain units of \(\Omega \mathrm{-m}\).coil: A region where toroidal current can flow with specified amplitude throughset_coil_currentsor via shape optimizationset_coil_regandset_isoflux
When defining the upper and lower shell segments we also pass noncontinuous=True to indicate that these regions are not toroidal continuous and thus the net toroidal current must be zero. Note that in this case the resistivity (eta) should be an effective resistivity that takes into account the toroidal coverage. See TokaMaker Example: Equilibria with toroidally non-continuous conductors in LTX for more information.
Define geometry for region boundaries
Once the region types and properties are defined we now define the geometry of the mesh using shapes and references to the defined regions.
- We add the limiter contour as a "polygon", referencing
plasmaas the region enclosed by the contour andairas the region outside the contour. - We add the upper shell as an "polygon" and the lower shell similarly by relfecting the contour. We also reference
airas the region outside the contour. - We add each of the sub-regions of the VV as "polygon"s and reference
airas the region outside the contour. - We add each of the 17 coils as "rectangles", which are defined by a center point (R,Z) along with a width (W) and height (H). We also reference
airas the region outside each region.
Plot topology
After defining the logical and physical topology we can now plot the curves within the definitions to double check everything is in the right place. In cases where curves appear to cross eachother (as with the OH1 and GREEN coils) one should zoom in to ensure no crossings exist. In this case we had to move the GREEN coils out by 1.27 cm to avoid an intersection with the OH1. Note a similar shift was also applied in the PSI-Tri model.
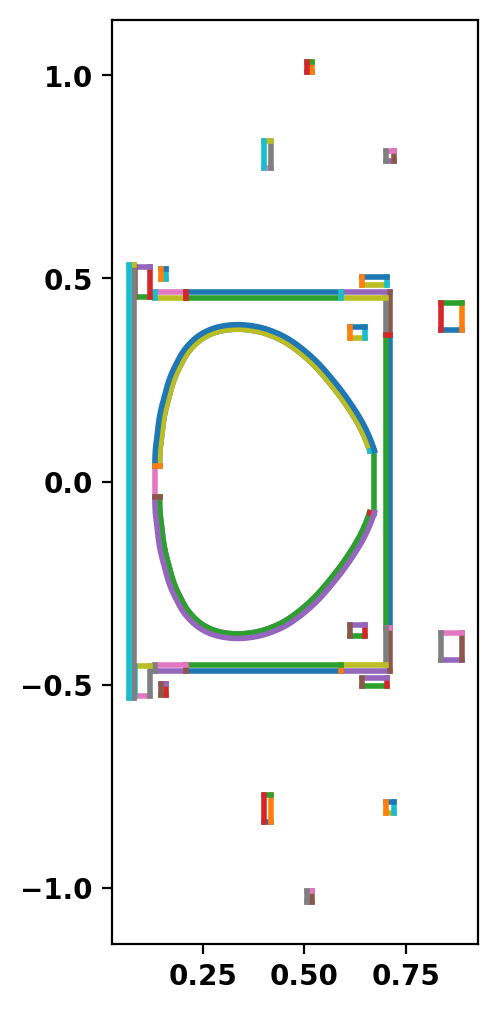
Create mesh
Now we generate the actual mesh using the build_mesh() method. Additionally, if coil and/or conductor regions are defined the get_coils() and get_conductors() methods should also be called to get descriptive dictionaries for later use in TokaMaker. This step may take a few moments as triangle generates the mesh.
Note that, as is common with unstructured meshes, the mesh is stored a list of points mesh_pts of size (np,2), a list of cells formed from three points each mesh_lc of size (nc,3), and an array providing a region id number for each cell mesh_reg of size (nc,), which is mapped to the names above using the coil_dict and cond_dict dictionaries.
Assembling regions: Warning: small feature (dl=1.59E-02) detected at point 397 (0.21, 0.466725) Warning: small feature (dl=1.59E-02) detected at point 399 (0.13335, 0.45085) Warning: small feature (dl=1.59E-02) detected at point 400 (0.59, 0.466725) Warning: small feature (dl=9.53E-03) detected at point 403 (0.7112, 0.36) Warning: small feature (dl=9.53E-03) detected at point 406 (0.7112, -0.36) Warning: small feature (dl=1.59E-02) detected at point 411 (0.59, -0.45085) Warning: small feature (dl=1.59E-02) detected at point 413 (0.21, -0.45085) Warning: small feature (dl=1.59E-02) detected at point 415 (0.13335, -0.45085) # of unique points = 590 # of unique segments = 108 Generating mesh: # of points = 3128 # of cells = 6114 # of regions = 28
Plot resulting regions and grid
We now plot the mesh by region to inspect proper generation.
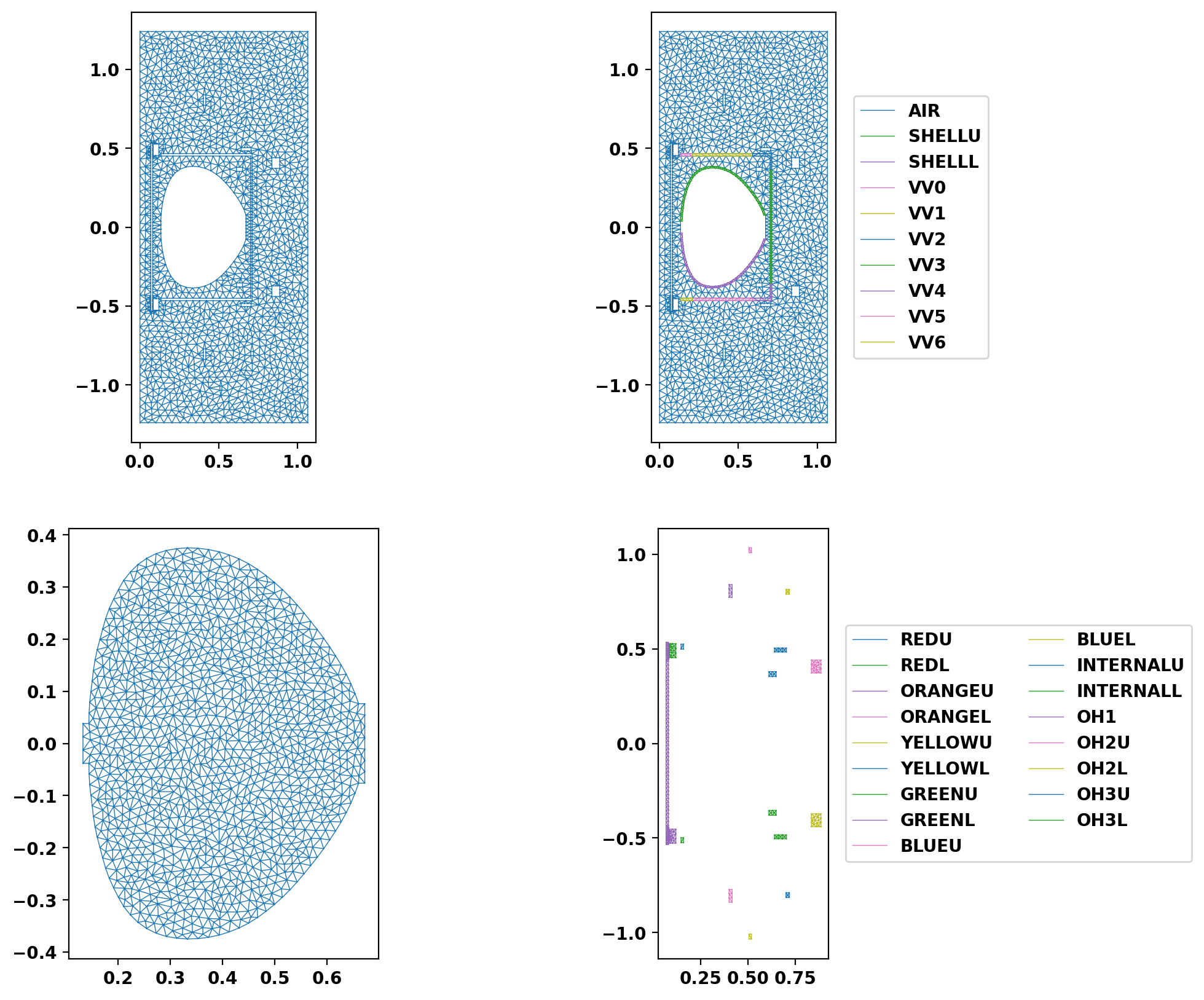
Save mesh for later use
As generation of the mesh often takes comparable, or longer, time compare to runs in TokaMaker it is useful to separate generation of the mesh into a different script as demonstrated here. The method save_gs_mesh() can be used to save the resulting information for later use. This is done using and an HDF5 file through the h5py library.